How to calculate the area of a certain class in a GeoTIFF file using NumPy
1 Introduction
Try these exercises first:
- How to read a GeoTIFF file as a NumPy array using GDAL
- Exercise: Plot a GeoTIFF file
- Exercise: Calculate the mean cell value of a GeoTIFF file
- How to extract cell values from a GeoTIFF file using NumPy
- How to classify a GeoTIFF file using NumPy
- How to detect changes between two GeoTIFF files using NumPy
- How to save a NumPy array as a GeoTIFF file using GDAL
In this exercise, we want to calculate the area of a certain class in a GeoTIFF file. Let’s calculate the area of waterbodies in an NLCD dataset. Use nlcd2016_clipped.tif.
2 Python code
from osgeo import gdal
import numpy as np
import matplotlib.pyplot as plt
def read_geotiff(filename):
ds = gdal.Open(filename)
band = ds.GetRasterBand(1)
arr = band.ReadAsArray()
return arr, ds
def write_geotiff(filename, arr, in_ds):
if arr.dtype == np.float32:
arr_type = gdal.GDT_Float32
else:
arr_type = gdal.GDT_Int32
driver = gdal.GetDriverByName("GTiff")
out_ds = driver.Create(filename, arr.shape[1], arr.shape[0], 1, arr_type)
out_ds.SetProjection(in_ds.GetProjection())
out_ds.SetGeoTransform(in_ds.GetGeoTransform())
band = out_ds.GetRasterBand(1)
band.WriteArray(arr)
band.FlushCache()
band.ComputeStatistics(False)
nlcd16_arr, nlcd16_ds = read_geotiff("nlcd2016_clipped.tif")
nlcd16_water_arr = np.where(nlcd16_arr == 11, 1, 0)
nlcd16_water_ncells = np.sum(nlcd16_arr == 11)
# https://gdal.org/tutorials/geotransforms_tut.html
dx = nlcd16_ds.GetGeoTransform()[1]
dy = -nlcd16_ds.GetGeoTransform()[5]
print("Total area of waterbodies is", nlcd16_water_ncells * dx * dy)
write_geotiff("nlcd16_water.tif", nlcd16_water_arr, nlcd16_ds)
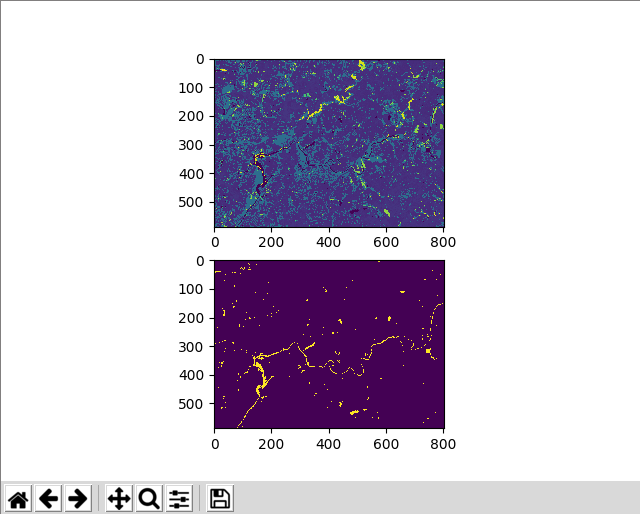
plt.subplot(211)
plt.imshow(nlcd16_arr)
plt.subplot(212)
plt.imshow(nlcd16_water_arr)
plt.show()3 Explanation
Import necessary modules and define the read and write functions.
from osgeo import gdal
import numpy as np
import matplotlib.pyplot as plt
def read_geotiff(filename):
ds = gdal.Open(filename)
band = ds.GetRasterBand(1)
arr = band.ReadAsArray()
return arr, ds
def write_geotiff(filename, arr, in_ds):
if arr.dtype == np.float32:
arr_type = gdal.GDT_Float32
else:
arr_type = gdal.GDT_Int32
driver = gdal.GetDriverByName("GTiff")
out_ds = driver.Create(filename, arr.shape[1], arr.shape[0], 1, arr_type)
out_ds.SetProjection(in_ds.GetProjection())
out_ds.SetGeoTransform(in_ds.GetGeoTransform())
band = out_ds.GetRasterBand(1)
band.WriteArray(arr)
band.FlushCache()
band.ComputeStatistics(False)Read the data into a NumPy array and get the GDAL dataset of the file. Remember, read_geotiff() returns two items.
nlcd16_arr, nlcd16_ds = read_geotiff("nlcd2016_clipped.tif")Based on the NLCD classification table, we extract open water cells only using the nlcd16_arr == 11 condition where 11 indicates the class of open water.
nlcd16_water_arr = np.where(nlcd16_arr == 11, 1, 0)Using the same condition nlcd16_arr == 11, count the number of open water cells. If a cell value is equal to 11, its value in the boolean array nlcd16_arr == 11 is True. If not, its value is False. The shape of this boolean array is the same as that of the original nlcd16_arr array. Now, using np.sum(), we add all the cell values in the boolean array. For this summation, True is equivalent to 1 and False to 0. In other words, we add 1 as many times as there are True cells and 0 as many times as there are False cells. However, since 0 does not really contribute to the summation result and 1 only contributes to it once, the result of this summation is the number of True cells. Which cells are True again? Yes, open water cells. Finally, we have the number of open water cells in the variable nlcd16_water_ncells.
nlcd16_water_ncells = np.sum(nlcd16_arr == 11)By referring to the geotransform tutorial, we extract the resolution of the NLCD dataset using the GetGeoTransform() function in the input dataset (nlcd16_ds). At index 1, we can find the x-resolution. The y-resolution is at index 5, but it is negative for a north-up image and we need to invert that (-nlcd16_ds.GetGeoTransform()[5] instead of just nlcd16_ds.GetGeoTransform()[5]).
# https://gdal.org/tutorials/geotransforms_tut.html
dx = nlcd16_ds.GetGeoTransform()[1]
dy = -nlcd16_ds.GetGeoTransform()[5]The area of each cell can be calculated by multiplying the x- and y-resolutions. Then, the total area of waterbodies is calculated by multiplying the number of waterbody cells and the cell area.
print("Total area of waterbodies is", nlcd16_water_ncells * dx * dy)Save the waterbodies array.
write_geotiff("nlcd16_water.tif", nlcd16_water_arr, nlcd16_ds)Finally, plot all the arrays.
plt.subplot(211)
plt.imshow(nlcd16_arr)
plt.subplot(212)
plt.imshow(nlcd16_water_arr)
plt.show()